Tutorial: Import IDs from SRA
After exporting your high-throughput sequencing data to SRA, SRA will attribute IDs to them. This tutorial explains how to use the script import_sra_id to import these SRA IDs and link them to your internal IDs. This allows you to keep track of published data within your own database.
To perform this linkage, import_sra_id uses two pieces of information:
- the number of spots (i.e. reads) sequenced,
- the columns
sample_refandreplicate_refadded as supplementary annotations during SRA export byexport_srascript. For example, see SRP189389 to find an example containing these columns.
Requirement
For this tutorial, it is expected that some data have been already exported to SRA. When you imported annotations following the Import your data tutorial, IDs were automatically attributed. But we attributed different IDs in our database. For the importing of SRA IDs to work, the local and SRA IDs need to be the same. To import our annotations:
-
Empty the seq tables. Execute
psql -U postgresto connect to PostgreSQL server and execute:TRUNCATE seq.project; TRUNCATE seq.sample; TRUNCATE seq.replicate; TRUNCATE seq.run;Exit Postgresql.
-
Import the annotations (SQL files are available here):
psql -U postgres < seq_project.sql psql -U postgres < seq_sample.sql psql -U postgres < seq_replicate.sql psql -U postgres < seq_run.sql
You can then check that you imported the data properly:
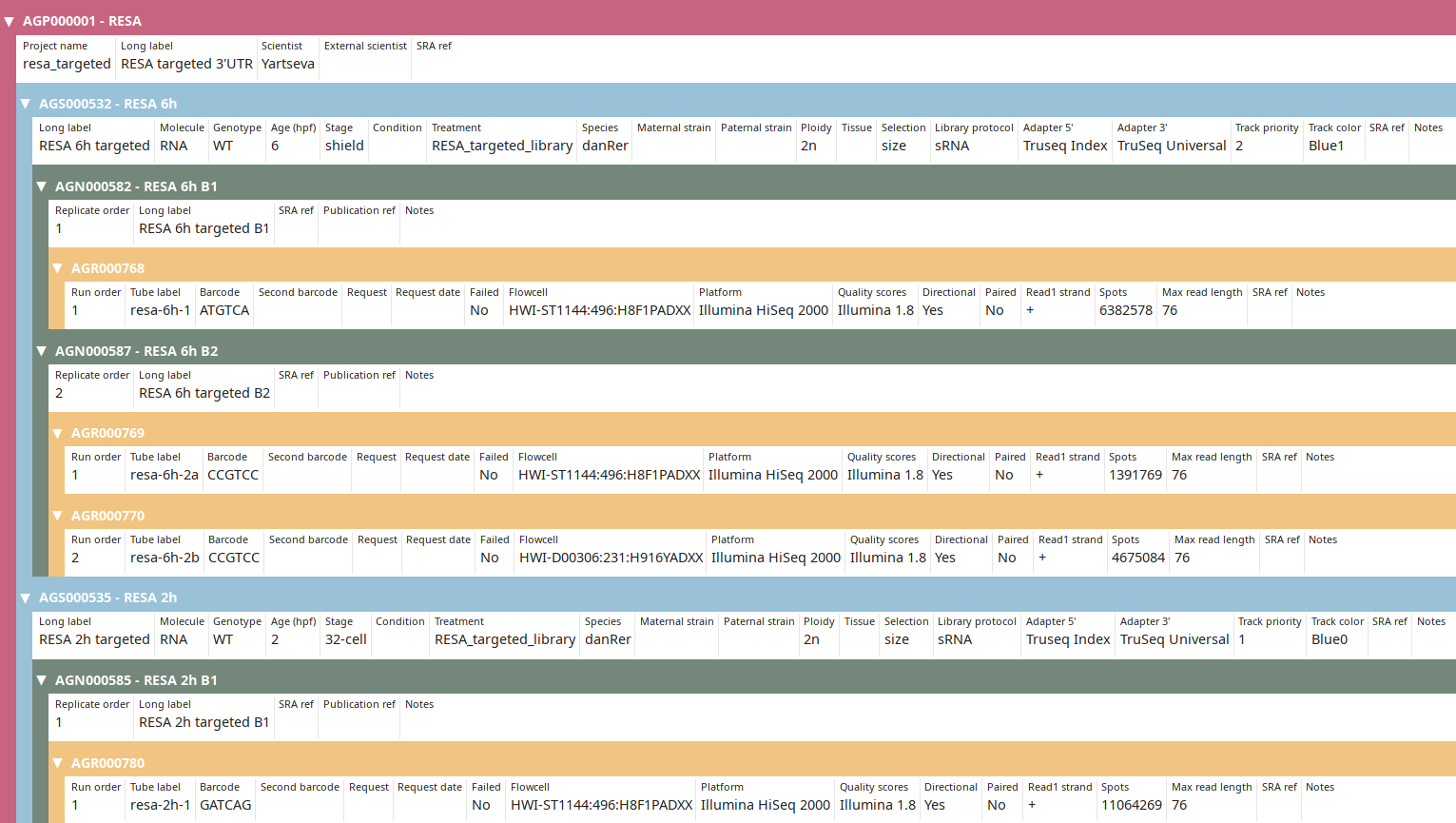
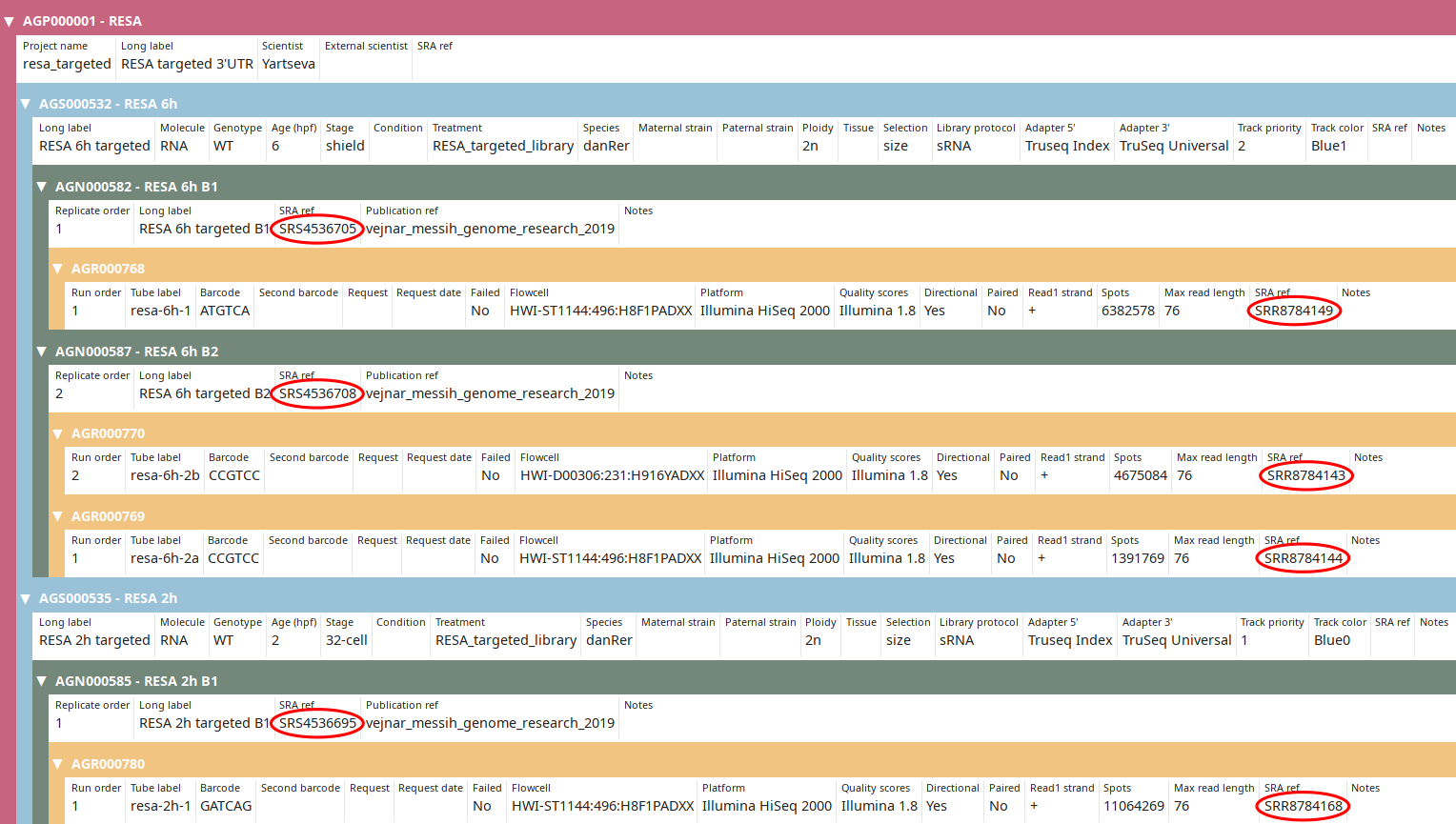
Observe that the IDs and the number of spots have been updated to new ones (compared to tutorial).
Create publication
Go to the Publications tab and click on the Add new Publication button to go the new publication form.
Use the following data:
| Column | Description |
|---|---|
| Publication (short internal name for publication) | vejnar_messih_genome_research_2019 |
| Title | Genome wide analysis of 3’-UTR sequence elements and proteins regulating mRNA stability during maternal-to-zygotic transition in zebrafish |
| Publication date | 2019-07-01 |
| Pubmed ID | 31227602 |
| SRA ref (comma separated list of SRP IDs) | SRP189512,SRP189389,SRP189499 |
…to fill the form:
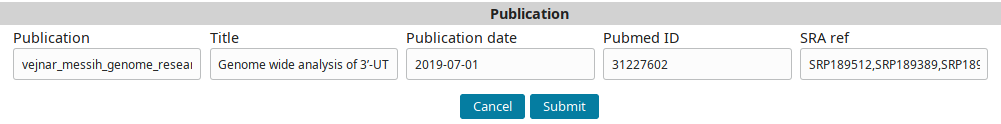
And submit to add the publication record.
Import
SRA IDs can then be imported using the --update option.
import_sra_id --update \
--publication_ref vejnar_messih_genome_research_2019 \
--dry | grep -v WARNING
import_sra_id won’t be able to find all the runs published within these SRA projects. It will display a warning for each unfound run. We suggest to first filter the numerous warnings. Run the command again the command without the | grep -v WARNING pipe to display these warnings.
This command returns:
Title: Genome wide analysis of 3’-UTR sequence elements and proteins regulating mRNA stability during maternal-to-zygotic transition in zebrafish
Loading SRP189512
> SRP189512
Loading SRP189389
> SRP189389
Replicate
> Local: AGN000585 RESA 2h B1
> SRA: RESA - WT 32c r2 B1 AGN000585 SRS4536695
> Run: SRR8784168
Replicate
> Local: AGN000582 RESA 6h B1
> SRA: RESA - WT 6h r2 B1 AGN000582 SRS4536705
> Run: SRR8784149
Replicate
> Local: AGN000587 RESA 6h B2
> SRA: RESA - WT 6h r2 B2 AGN000587 SRS4536708
> Run: SRR8784143
Replicate
> Local: AGN000587 RESA 6h B2
> SRA: RESA - WT 6h r2 B2 AGN000587 SRS4536708
> Run: SRR8784144
Loading SRP189499
> SRP189499
...
Once you got this output, execute the command again without the --dry option:
import_sra_id --update \
--publication_ref vejnar_messih_genome_research_2019 | grep -v WARNING
Now the SRA IDs should be imported as you can see in the tree view: