Tutorial: Import data from SRA
The import_sra script imports data and sample annotations from publicly available publicly available high-throughput sequencing data from SRA. It will:
- Downloads the FASTQ files,
- Format the data to comply with a common format. This includes by default renaming files, compressing the FASTQ files with a more efficient algorithm than Gzip, and stripping read names.
- Input the sequencing run(s) into the database.
Download
Imported data were published in Yartseva et al under accession SRP090954.
import_sra --project SRP090954 \
--runs SRR4375304,SRR4375305 \
--path_seq_run /data/seq/by_run \
--path_seq_prepared /data/seq/prepared \
--dump_sra \
--db_import \
--create_links
The --dump_sra option downloads and gets FASTQ files, --db_import imports samples and runs into the database, while --create_links creates the symbolic links to /data/seq/by_run.
--project option is used, all runs will be imported. If only a few runs are needed, use the --runs option.
This command returns:
Downloading infos # 0 ID 3268484
Downloading infos # 1 ID 3268483
Downloading infos # 2 ID 3268482
Downloading infos # 3 ID 3268481
Downloading infos # 4 ID 3268480
Downloading infos # 5 ID 3268479
Downloading infos # 6 ID 3268478
Downloading infos # 7 ID 3268477
Downloading infos # 8 ID 3268463
Downloading infos # 9 ID 3268411
Downloading infos # 10 ID 3268394
Downloading infos # 11 ID 3268393
Downloading infos # 12 ID 3268378
Downloading infos # 13 ID 3268358
Downloading infos # 14 ID 3268357
Downloading infos # 15 ID 3268356
Downloading infos # 16 ID 3268355
Downloading infos # 17 ID 3268354
Downloading infos # 18 ID 3268353
Downloading infos # 19 ID 3268352
Downloading infos # 20 ID 3268351
Downloading infos # 21 ID 3268350
Downloading infos # 22 ID 3268349
RESA identifies mRNA regulatory sequences with high resolution
SRS1732690 RESA-Seq - WT 8h pA r3 B1
\_SRR4375304
SRS1732691 RESA-Seq - WT 8h pA r3 B2
\_SRR4375305
Import to DB
Dump from SRA
Download and dump SRR4375304
join :|-------------------------------------------------- 100.00%
concat :|-------------------------------------------------- 100.00%
spots read : 5,896,053
reads read : 11,792,106
reads written : 11,792,106
SRR4375304_R1.fastq : 20.71% (1036594224 => 214688034 bytes, SRR4375304_R1.fastq.zst)
SRR4375304_R2.fastq : 20.44% (1036594224 => 211867665 bytes, SRR4375304_R2.fastq.zst)
Download and dump SRR4375305
join :|-------------------------------------------------- 100.00%
concat :|-------------------------------------------------- 100.00%
spots read : 8,845,249
reads read : 17,690,498
reads written : 17,690,498
SRR4375305_R1.fastq : 20.35% (1555652720 => 316532334 bytes, SRR4375305_R1.fastq.zst)
SRR4375305_R2.fastq : 20.83% (1555652720 => 324036832 bytes, SRR4375305_R2.fastq.zst)
Imported runs from the SRP090954 project are large. For a quick test of import_sra, import SRR1761155 from SRP052298.
import_sra --project SRP052298 \
--runs SRR1761155 \
--path_seq_run /data/seq/by_run \
--path_seq_prepared /data/seq/prepared \
--dump_sra \
--db_import \
--create_links
Result
Files
The import_sra script creates the following files including the data files (in prepared folder as files are converted from SRA to FASTQ files) and symbolic links:
/data/seq
├── [4.0K] by_run
│ ├── [4.0K] SRR4375304
│ │ ├── [ 48] SRR4375304_R1.fastq.zst -> ../../prepared/SRP090954/SRR4375304_R1.fastq.zst
│ │ └── [ 48] SRR4375304_R2.fastq.zst -> ../../prepared/SRP090954/SRR4375304_R2.fastq.zst
│ └── [4.0K] SRR4375305
│ ├── [ 48] SRR4375305_R1.fastq.zst -> ../../prepared/SRP090954/SRR4375305_R1.fastq.zst
│ └── [ 48] SRR4375305_R2.fastq.zst -> ../../prepared/SRP090954/SRR4375305_R2.fastq.zst
└── [4.0K] prepared
└── [4.0K] SRP090954
├── [205M] SRR4375304_R1.fastq.zst
├── [202M] SRR4375304_R2.fastq.zst
├── [302M] SRR4375305_R1.fastq.zst
└── [309M] SRR4375305_R2.fastq.zst
Annotations
The import_sra script imports the sequencing run(s) into the database. Use the Tree view to display the new project:
- SRPxxx and SRRxxx IDs are conserved and imported as-is from SRA.
- SISxxx (SRA Import Sample) and (SRA Import N replicate) are created as no IDs in SRA correspond to LabxDB definition of samples and replicates and not all data providers interpret the definition of SRA samples the same way.
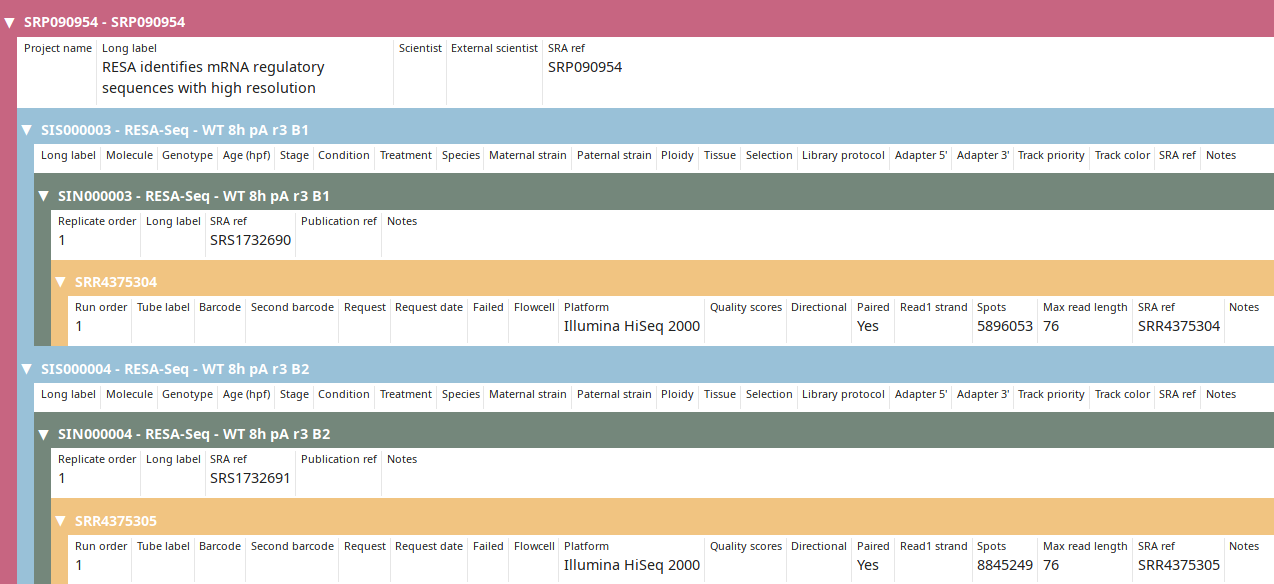
After annotations are imported from SRA, project and sample can be further annotated manually.