Tutorial: Export your data to SRA
The export_sra script prepares your sample annotations to easily export them into SRA. It export annotations into tabulated files following SRA expected format. The associated script export_sra_fastq prepared the FASTQ files, ready to be uploaded into SRA. This process is required when publishing your data.
Requirement
For this tutorial, it is expected that the tutorial to import your own data has been followed. To skip this tutorial and start at the same stage,
-
Empty the seq tables. Execute
psql -U postgresto connect to PostgreSQL server and execute:TRUNCATE seq.project; TRUNCATE seq.sample; TRUNCATE seq.replicate; TRUNCATE seq.run;Exit PostgreSQL.
-
Import the annotations (SQL files are available here):
psql -U postgres < seq_project.sql psql -U postgres < seq_sample.sql psql -U postgres < seq_replicate.sql psql -U postgres < seq_run.sql
Export annotations
To export the three replicates from the RESA project, execute:
export_sra --replicates AGN000001,AGN000002,AGN000003
The labels used internally in your database might be different than the appropriates labels for SRA. To apply filters on labels, add the --path_label_filters option. Filters are submitted in a CSV file: the first column is the string to be replaced by the string defined in the second column. For example:
RESA,RESA (RNA-element selection assay)
To use the filter (saved in filters.csv), execute:
export_sra --replicates AGN000001,AGN000002,AGN000003 \
--path_label_filters filters.csv
export_sra will create:
sra_samples.tsvwith the sample annotationssra_data.tsvwith a list of exported FASTQ filessra_exported.tsvwith already SRA exported samples files
How to use these files is explained in the next section.
Export FASTQ files
To prepare the corresponding FASTQ files using the data.json created in the previous step, execute:
export_sra_fastq --path_seq_raw '/data/seq/raw' \
--path_seq_run '/data/seq/by_run' \
--path_data data.json
The following files will be created:
├── AGR000001_R1.fastq.gz
├── AGR000002_R1.fastq.gz
├── AGR000003_R1.fastq.gz
├── AGR000004_R1.fastq.gz
├── data.json
├── sra_data.tsv
├── sra_exported.tsv
└── sra_samples.tsv
Submit to SRA
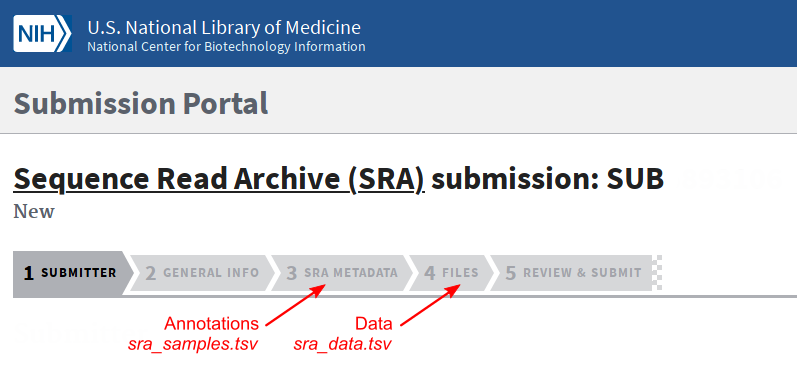
To submit samples to SRA, login to the Submission Portal and go to the Sequence Read Archive (SRA).
Then click on the New submission button to start a new submission:
Fill the different forms. Use the sra_samples.tsv and sra_data.tsv files in the appropriate tabs.
At the end of the submission, you can upload the FASTQ files.