Features
Databases
LabxDB comes with ready-to-use databases. Data is stored in PostgreSQL. LabxDB provides web-browser GUI and a programmatic API for intregration into pipelines.
Research
Research-oriented databases for plasmid collections, cell lines, fly or fish mutant lines are ready-to-use.
HT sequencing
LabxDB is ready for annotating high-throughput sequencing samples and interacting with public resources such as NCBI SRA.
Lab management
LabxDB facilitates laboratories management such as managing purchasing, easily centralizing information and keeping tracks of lab history.
Modular
LabxDB provides generic reusable and extensible blocks to create new databases.
Structured data
LabxDB databases can store hierarchical information.
Customizable
Specific functionalities can easily be added to GUI and backend.
Databases
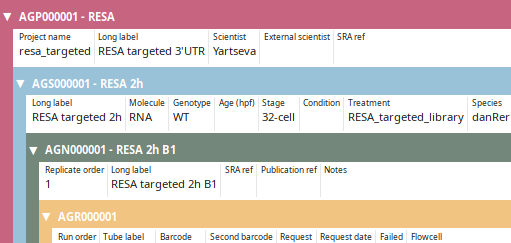
LabxDB seq
LabxDB for high-throughput sequencing (HTS) annotation. LabxDB seq organizes sample annotations using a structure of nested levels: from the top-level projects to the bottom level sequencing runs. LabxDB provides forms to easily create sample structure, to annotate and view (as captured by the image on the left) all levels at the same time. LabxDB seq comes with scripts to import HTS data from local sequencing facilities or public resources.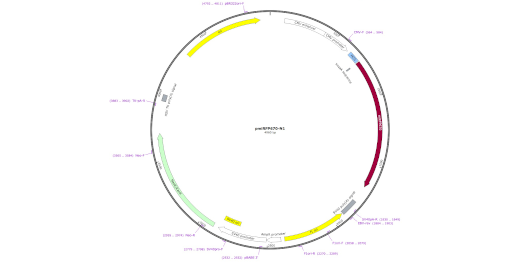
LabxDB plasmid
Storage and management for a collection of plasmids, including the full plasmid sequence and a picture of the plasmid map.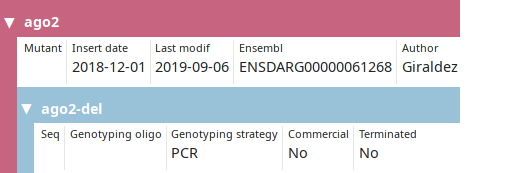
LabxDB mutant line
Collection of mutant cell or organism. A nested "allele" level allows the user to store multiple alleles for each mutant if necessary.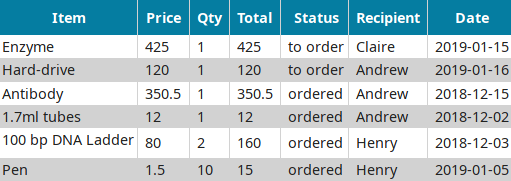
LabxDB order
Centralizing purchases within a lab while tracking spending.Publication
LabxDB: versatile databases for genomic sequencing and lab management
Charles E. Vejnar and Antonio J. Giraldez
Bioinformatics, Volume 36, Issue 16, August 2020
https://doi.org/10.1093/bioinformatics/btaa557
Charles E. Vejnar and Antonio J. Giraldez
Bioinformatics, Volume 36, Issue 16, August 2020
https://doi.org/10.1093/bioinformatics/btaa557